I have been playing around with the Python language for a month or two, and really enjoy it. Specifically, I have been using Matthew Russell’s book Mining the Social Web (no financial relation here, just a happy customer) to find out how to do my own custom social network analysis. I’ve really only dipped my toes in it, but even the simple stuff can yield great insights. (Perhaps that’s the greatest insight I’m getting from this book!)
At any rate, I basically compiled all the material from Chapter 1 into one program, and searched for the term #pharma. I came up with the following:
Search term: #pharma
Total words: 7685
Total unique words: 3142
Lexical diversity: 0.408848
Avg words per tweet: 15.370000
The 50 most frequent words are
[u'#pharma', u'to', u'-', u'RT', u'#Pharma', u'in', u'for', u'the', u'and', u'of
', u':', u'a', u'on', u'&', u'#Job', u'is', u'#jobs', u'Pharma', u'#health',
u'#healthinnovations', u'\u2013', u'The', u'at', u'#hcmktg', u'Pfizer', u'from'
, u'US', u'with', u'#biotech', u'#nhs', u'de', u'#drug', u'drug', u'http://bit.ly/PHARMA', u'this', u'#advertising', u'...', u'FDA', u'#Roche', u'#healthjobs',
u'an', u'Medical', u'that', u'AdPharm', u'them', u'you', u'#medicament', u'Big',
u'ouch', u'post']
The 50 least frequent words are
[u'verteidigen', u'video', u'videos,', u'vids', u'viral', u'visiting', u'vom', u
'waiting', u'wave', u'way:', u'weakest', u'website.', u'week', u'weekend,', u'we
ekly', u'werden', u'whether', u'wholesaler', u'willingness', u'winning...', u'wi
sh', u'work?', u'workin...', u'working', u'works', u'works:', u'world', u'worldw
ide-', u'write', u'wundern', u'www.adecco.de/offic...', u'year', u'years,', u'ye
t?', u"you'll", u'you?', u'youtube', u'you\u2019re', u'zone', u'zu', u'zur', u'{
patients}!?', u'\x96', u'\xa360-80K', u'\xbb', u'\xc0', u'\xe9cart\xe9s', u'\xe9
t\xe9', u'\xe9t\xe9...', u'\xfcber']
Number of users involved in RT relationships for #pharma: 158
Number of RT relationships: 108
Average number of RTs per user: 1.462963
Number of subgraphs: 52
(The term u'<stuff>' above simply means that the words in quotes are stored in unicode, which you can safely ignore here.)
As for graphing the retweet relationships, I came up with the following:
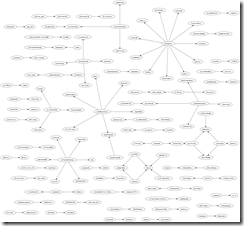
So whom should you follow if you want pharma news? Probably you can start with those tweeters at the centers of wheels with a lot of spokes. What are #pharma tweeters concerned about these days? Jobs, jobs, jobs, and maybe some innovations.
Update: I checked the picture and saw that it was downsized into illegibility. So I uploaded an svg version.

